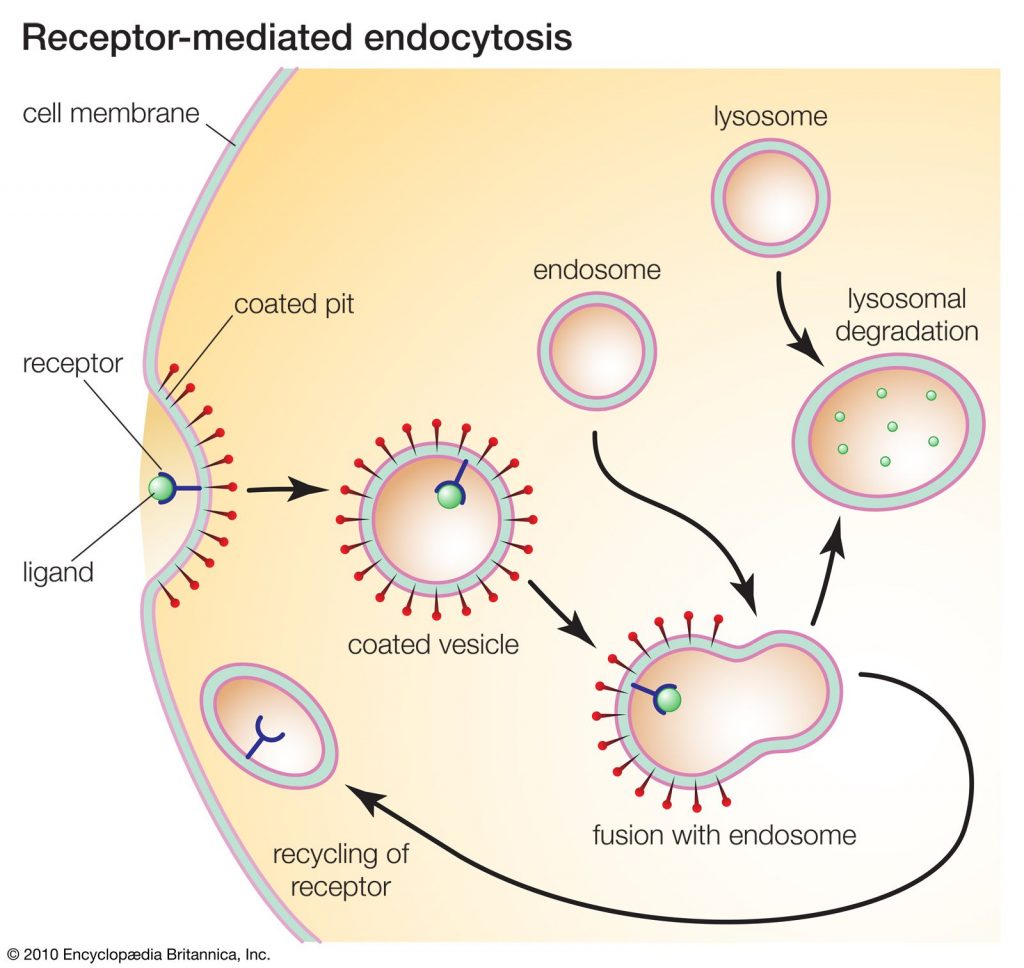
Definition of receptors Receptors are proteins, usually, cell surface receptors, that bind ligands and elicit responses in the immune system, including cytokine receptors, growth factor receptors, and the Fc receptor. Receptors can be found on various immune cells such as B cells, T cells, NK cells, monocytes, and stem cells. A molecule that binds to Read More
Cusabio N-terminal 6xHis-GST-tagged Recombinant
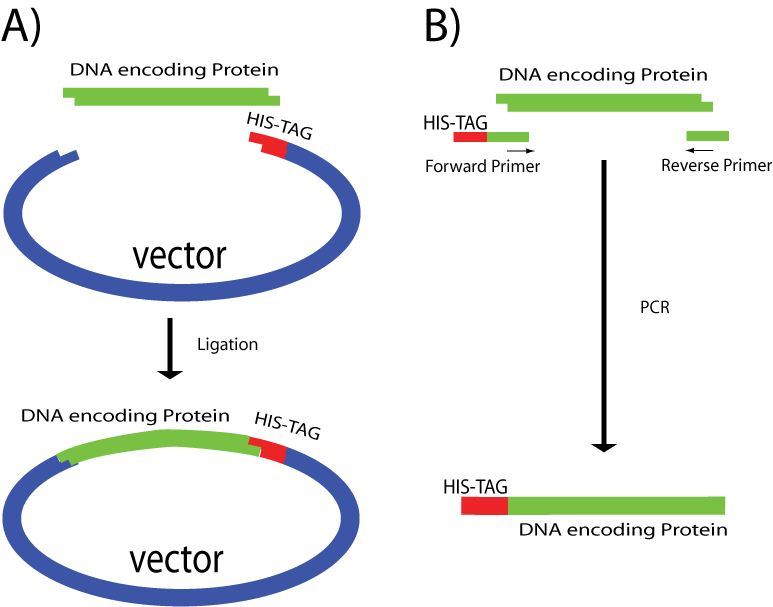
The GST label Protein purification with affinity tags, such as glutathione S-transferase (GST), histidine (HIS) and other affinity tags, allows the purification of proteins with both known and unknown biochemical properties. Therefore, this methodology has become a widely used research tool to determine the biological function of uncharacterized proteins. N-terminal 6xHis-GST-tagged Recombinant is a 211 Read More
Cusabio Biochemicals Recombinants
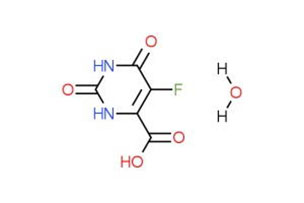
Abstract Human dimethylglycine dehydrogenase (hDMGDH) is a flavin adenine dinucleotide (FAD) and tetrahydrofolate (THF)-dependent mitochondrial matrix enzyme that is involved in choline degradation, one-carbon metabolism, and electron transfer to the respiratory chain. . The rare natural variant H109R causes a deficiency of dimethylglycine dehydrogenase leading to increased concentrations of dimethylglycine in the blood and urine. Read More
Unusual aetiology of respiratory compromise in a patient with AIDS
A 36-year-old African American man with no medical historical past offered with a current historical past of cough and dyspnoea. Initial chest imaging revealed diffuse bilateral lung infiltrates. A subsequent HIV check resulted constructive, and he was presumptively identified with AIDS, later confirmed by a CD4 of 88 cells/mm3 Empiric remedy with trimethoprim-sulfamethoxazole was initiated for Read More
Exploration of a Sequential Gp140-Gp145 Immunization Regimen with Heterologous Envs to Induce a Protective Cross-Reactive HIV Neutralizing Antibody Response In Non-human Primates
Raising a heterologous tier 2 neutralizing antibody (nAb) response stays a daunting job for HIV vaccine growth. In this research, we explored the utility of numerous HIV-1 envelope (Env) immunogens in a sequential immunization scheme as a answer to this job. This exploration stemmed from the rationale that gp145, a membrane-bound truncation kind of HIV Read More
Characterization of humoral and SARS-CoV-2 specific T cell responses in people living with HIV
There is an pressing want to grasp the character of immune responses in opposition to SARS-CoV-2, to tell risk-mitigation methods for people living with HIV (PLWH). We present that almost all of PLWH, managed on ART, mount a practical adaptive immune response to SARS-CoV-2. Humoral and SARS-CoV-2-specific T cell responses are comparable between HIV-positive and Read More
Genetic characterization of a novel HIV-1 second-generation recombinant form (CRF01_AE/07_BC) identified in Yan`an city, China
Due to co-epidemic of CRF01_AE and CRF07_BC in China, growing numbers of the second-generation recombinants between them have been identified particularly amongst sexual inhabitants. Here, we identified an distinctive CRF01_AE/CRF07_BC recombinants from a male HIV-1 postive particular person (18YA004) contaminated by heterosexual contact in Yan`an metropolis, Shaanxi province. The close to full-length genome analyses confirmed Read More
Phylogenetic analysis of HIV-1 archived DNA in blood and gut-associated lymphoid tissue in two patients under antiretroviral therapy
One of the approaches to remedy human immunodeficiency virus (HIV) is the use of therapeutic vaccination. We have launched the Provir/Latitude 45 research to determine conserved CTL epitopes in archived HIV-1 DNA based on the HLA class I alleles in aviremic patients under antiretroviral therapy (ART). A HIV-1 polypeptidic therapeutic vaccine based mostly on viral Read More
Minimizing the Impact of the Triple Burden of COVID-19, Tuberculosis and HIV on Health Services in sub-Saharan Africa
In this angle, we talk about the affect of COVID-19 on tuberculosis (TB)/HIV well being companies and approaches to mitigating the rising burden of these three colliding epidemics in sub-Saharan Africa (SSA). SSA nations bear considerably excessive proportions of TB and HIV instances reported worldwide, in comparison with nations in the West. Whilst COVID-19 epidemiology Read More
Portable Smart-Space Research Interface to Predetermine Environment Acoustics for Cochlear implant and Hearing aid users with CCi-MOBILE
Internet of issues (IoT) in healthcare, has effi-ciently accelerated medical monitoring and evaluation by the real-time evaluation of collected knowledge. Hence, to assist the hearing-impaired group with higher calibrations to their medical processors and listening to aids, a conveyable sensible house interface – AURIS has been developed by the Cochlear Implant Processing Lab (CILab) at Read More